TrialAI
A proof-of-concept platform exploring how Large Language Models can accelerate and improve clinical research
Overview
TrialAI was created to explore how large language models could support clinical trial researchers in synthesizing, comparing, and designing more efficient protocols. The project examined workflows in which AI could assist in early trial design tasks without compromising scientific rigor or regulatory compliance.
The User
Actions & Motivations
- Designs and implements clinical trials with multidisciplinary teams.
- Ensures compliance with ethical and regulatory standards.
- Driven to improve patient outcomes through rigorous, efficient science.
- Seeks tools that enhance collaboration and reduce cognitive load.
Pains
- Balancing comprehensive criteria design with time pressure.
- Managing heavy workloads across multiple trials.
- Maintaining clear communication in distributed teams.
Additional Research Persona Insights
We also interviewed data analysts and medical scientists from the pharmaceutical and life sciences sectors. They shared complementary challenges around data interpretation, NLP application, and clinical data extraction.
- Goals: Accelerate drug discovery through improved data analysis and automation.
- Skills: Medical advancements, data analytics, and NLP for clinical datasets.
- Needs: Reliable biomedical NLP tools and AI systems to extract insights from EHRs.
- Pain Points: Limited time for deep analysis, rapidly evolving technologies, and uncertain research funding landscapes.
Conceptual Architecture
The architecture behind TrialAI connects backend clinical trial registries with an AI-assisted front-end interface. Structured data from sources like Citeline and ClinicalTrials.gov flows into a generative layer that powers intelligent search, summarization, and inclusion/exclusion criteria generation.
Data flow from backend registries to AI-powered front-end workflows
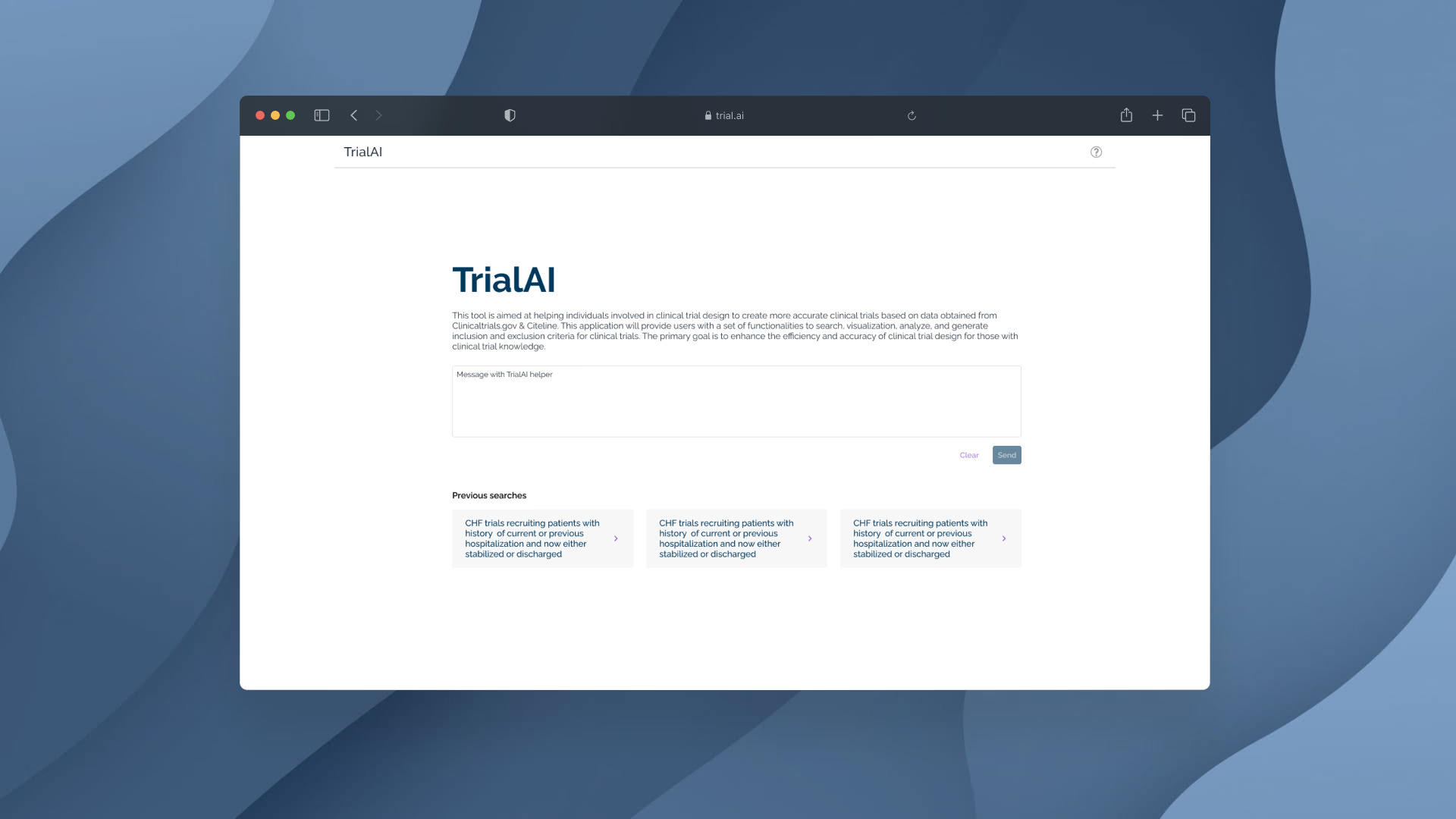
Prototype
The high-fidelity prototype illustrated how clinical researchers could interact conversationally with TrialAI to query and refine trial parameters. The chat interface provided a seamless bridge between data exploration and actionable design recommendations.
View PrototypeConclusion
Through this research and prototyping effort, TrialAI demonstrated how LLMs can reduce friction in trial design — supporting tasks such as literature review, protocol drafting, and eligibility criteria generation. The findings inform a roadmap toward ethical, explainable AI integration in biomedical research tools.